Welcome to riboCIRC v2.0
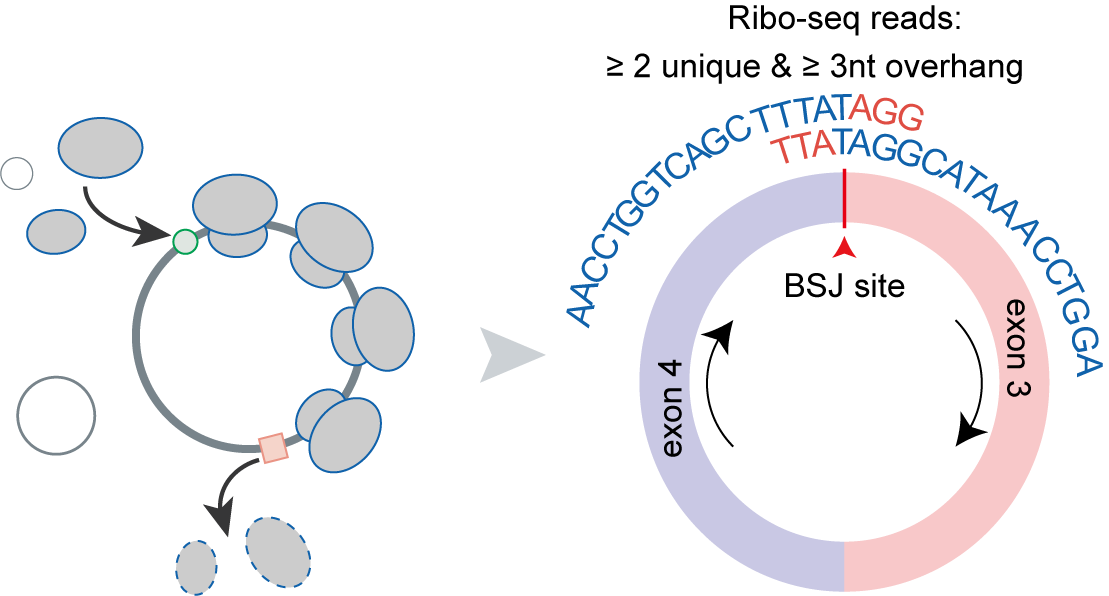
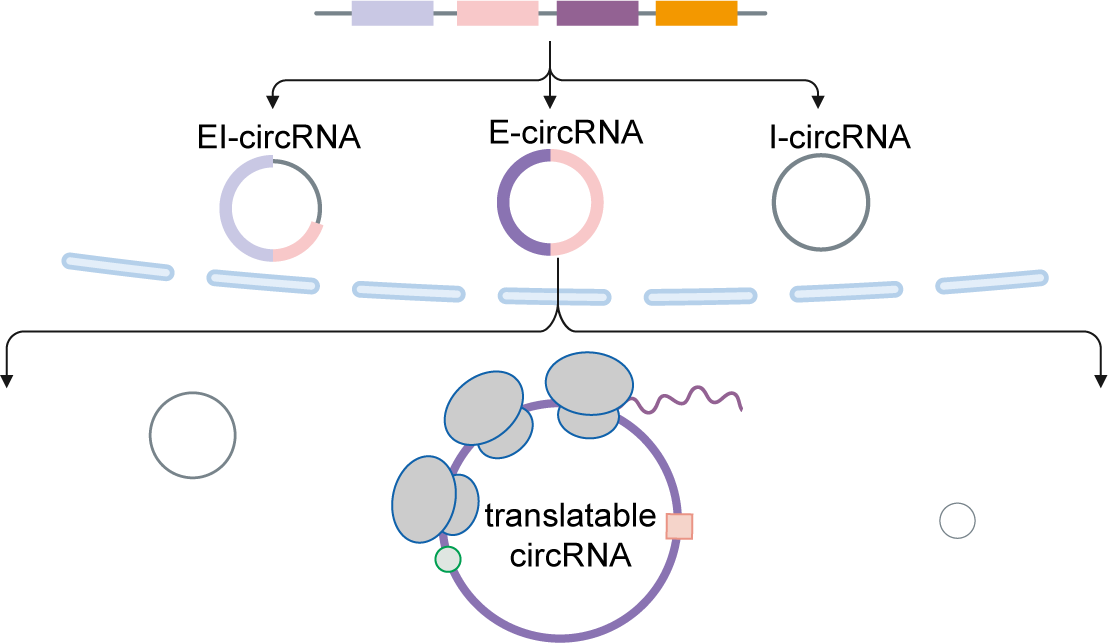
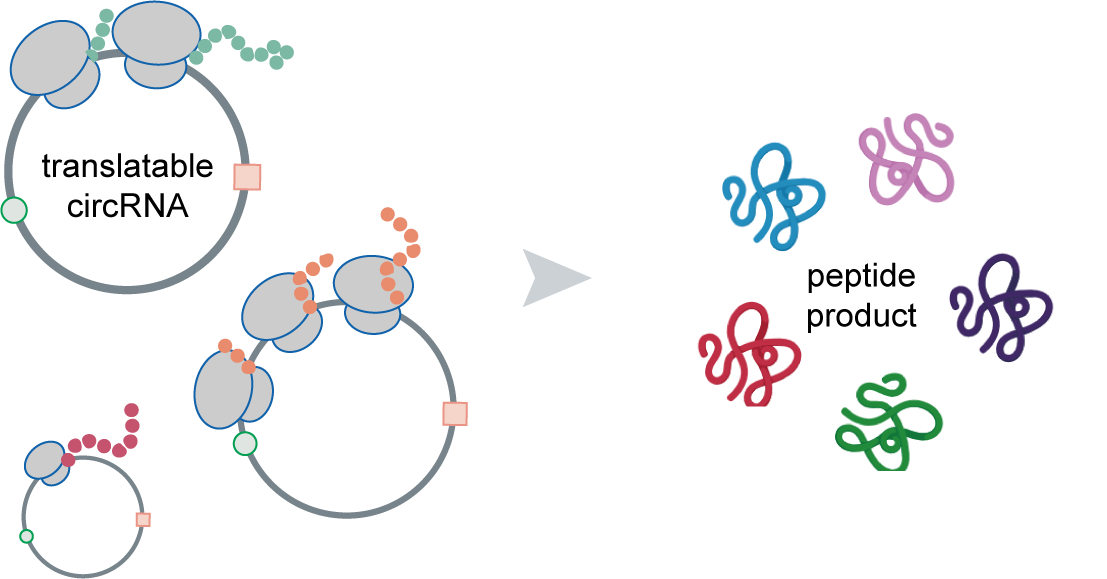
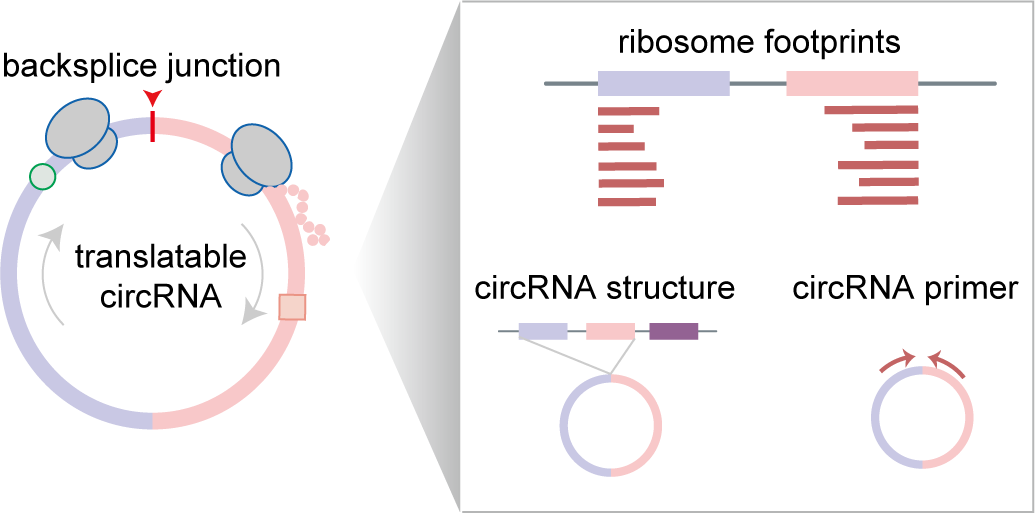
The translation of circular RNAs (circRNAs) is a rapidly expanding field, captivating researchers due to the crucial roles played by their encoded functional peptides in gene expression regulation. Despite this growing interest, the full landscape of circRNA translation remains largely unexplored. To address this knowledge gap, we initially developed riboCIRC, a translatome data-oriented multi-species database of translatable circRNAs (human, mouse, rat, worm, fly and zebrafish). Building upon its success, we have now launched riboCIRC v2.0, offering an even more comprehensive and insightful platform designed to illuminate the intricacies of circRNA translation. riboCIRC v2.0 provides: (1) A comprehensive repository of computationally predicted ribosome-associated circRNAs (ribo-circRNAs). This enhanced resource integrates diverse evidence supporting their translation, including BSJ-spanning ORF, IRES element, m6A modification, RBP binding, RNA editing site, and mass spectrometric proofs. (2) A meticulously curated collection of literature-reported translatable circRNAs. This ensures that experimentally validated instances of circRNA translation are readily accessible. (3) A robust evaluation of ribo-circRNA conservation across various species. This feature allows for the identification of evolutionarily conserved and potentially functionally significant translatable circRNAs. (4) A systematic de novo annotation of putative circRNA-encoded peptides. For each predicted peptide, we provide detailed information on its sequence, predicted secondary and tertiary structure, and potential biological function. (5) An intuitive genome browser. This tool enables users to visualize the context-specific ribosome footprints on circRNAs, offering direct insights into their translational activity across different conditions or cell types.
The database currently includes
News
-
25 Jun 2025
riboCIRC v2.0 is released.
-
8 Mar 2021
riboCIRC is published in Genome Biology.
-
15 Aug 2020
riboCIRC v1.0 is released.
How to cite
-
1. Li et al. riboCIRC: a comprehensive database of translatable circRNAs. Genome Biology. 2021 Mar 8;22(1):79.
-
2. Xu et al. riboCIRC v2.0: an expanded resource for translatable circRNAs. Nucleic Acids Research. 2025; 2025 Oct 21:gkaf1022.
Contact
- For questions or support, please contact us:
-

Correspondence:Hongwei Wang bioccwhw@126.com
-

Technical assistance:Mingzhe Xie xiemzh6@gmail.com